Directed evolution
flickr - Gaetan Lee
With the genes we proposed for development in cells, it is likely that we end up with a genetic circuit which is not working for what we intended, or even non-operational. Even in the case of a simple inverter, some characteristics we designed for the circuit are difficult to be ensured. This inverter may simply be not working for any input levels, or working but in undesired levels.
Laborious work could be done to fine tune the properties described above to yield a functional circuit as we intended, however even for this relatively straightforward example, this process is not trivial given that the properties of this circuit is determined by many complex biological phenomena. Especially when under the circumstances that the construction of more complex circuits would require more logic gates or analogue components, this would certainly become a serious problem.
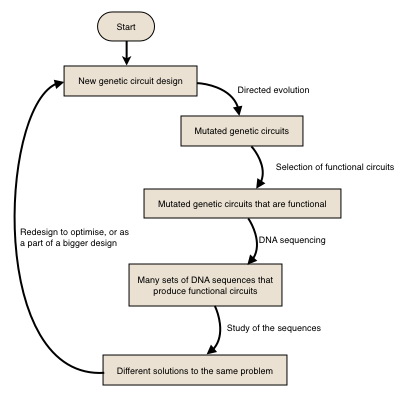
This is when directed evolution plays its part. It allows evolution, which is a rather slow process, to quickly occur in laboratory environment. It is a method for allowing random mutation of genes and selection of those with wanted performances, in order to evolve proteins in a way we wished. Usually, some of the evolved proteins will exhibit characteristics we wanted which we will discuss in detail.
It utilises a PCR process which is more error-prone, by introducing specific kind of divalent cations such as Mn2+, to rapidly increase the size of existing DNA library. Then we may identify and isolate mutants with required behaviours. After this, we allow mutants to replicate, and sequence their DNA molecules to understand the characteristics of mutations occurred. Finally, with our optimised and better studied genes, we could prepare and record their sequences for contributing as a component of future developments of more complex genetic circuits or networks, as illustrated in the flow chart below.
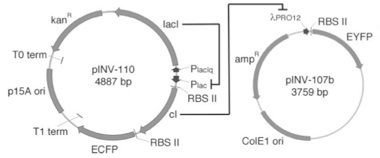
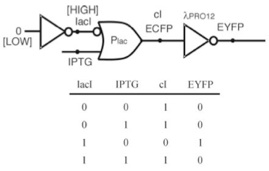
Let us consider a simple circuit (see figure below on the left) proposed by researchers from California Institute of Technology and Princeton University in 2002.1 This device functions similarly to the inverter we exemplified previously. In plasmid pNlV-110, without the presence of the input, lacI represses the promoter Plac, as a consequence the gene expression of cI is prevented. In plasmid pNlV-107b, cI if present in turn represses the λPRO12 promoter, and also the gene expression of EYFP (enhanced yellow fluorescent protein), which is a protein for producing fluorescence for easy detection and functions as the output from the circuit. Our input, IPTG (Isopropyl β-D-thiogalactoside) is an inducer which induces lacI. If present, it allows the production of cI, prevents expression of EYFP; if absent then the exact opposite would occur and EYFP could be expressed. This is analogous to the digital circuit shown below to the right.
When the circuit was first tested, it was a non-working device - no matter what level of the input IPTG is, it never produces the output. This is because under any conditions the cI was always adequate to repress the λPRO12 promoter, or in the language of digital electronics, the output potential from the Plac OR gate to represent ‘low’ is not small enough for the λPRO12 NOT gate to register as a ‘low’ input, always regarding the input to it as ‘high’, therefore the output is always ‘low’.
However after going through the process of directed evolution using error-prone PCR, in the absence of IPTG, they discovered 50% of mutants exhibits fluorescence,which is part of the desired behaviour (low input maps to high output). Then these mutants were transferred to a plate containing a desired concentration (800 μmolL-1) of input IPTG, they found 5% to 10% of mutants were non-fluorescent in this environment.
DNA sequencing was then used, it unveils many possible approaches to the original problem, such as mutations changed binding affinity between the repressor and operator, or the strength of RBS, which could affect transcription and translation efficiency respectively.
References
1 Yokobayashi, Y., Weiss, R.: Directed evolution of a genetic circuit (2002) (Including two figures below)