DNA sequencing
flickr - Shaury
One particularly essential task performed by genetic engineers is sequencing of the DNA molecules. This is a process which determines the sequence of nucleotides in a DNA molecule. Recall what we know about DNA, simply speaking, DNA sequencing is to deduce the order of A, T, C and G in a DNA molecule.
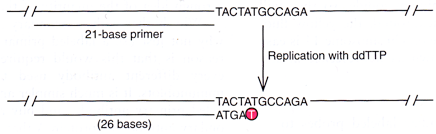
Similar to the replication of genes discussed in the previous section, it also utilises PCR to produce DNA fragments, and the first two steps are the same. However the difference between growing genes of interest and this method is that chemicals called dideoxynucleotides are present in the process of elongation. While the other principles remain almost identical to the PCR in the third step of elongation, in this case, dideoxynucleotides which are with the same function of nucleotides, can be used to form individual pieces at the end of a strand being elongated, however once it is added to the strand, the strand can no longer be extended. We can visualise this by again the analogy we used to discuss the elongation of primer. You may notice one thing is missing from the zipper, that is the top stop which stops the slider from moving further so it won’t fall apart from the zipper. The same applies in this case, dideoxynucleotides stop the polymerase from moving along the rest of the template strand, thus the polymerase will simply be detached from them. (see the figure below, the one labeled in a circle is the dideoxynucleotide derived from nucleobase T.)
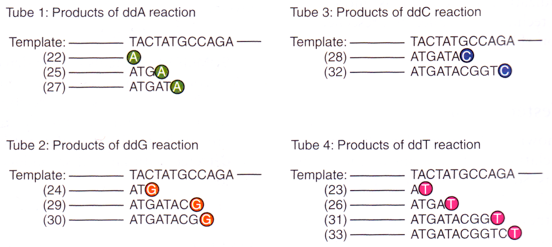
After dideoxynucleotides terminate elongation of the strand being extended, we will have segments ending at various positions of any of the four different bases of the template strand. If these three steps are allowed to be repeated many times for all different types of dideoxynucleotides, we will have all DNA molecules from smaller ones to bigger ones that are segments starting from the position of the primer and stopping at any possible bases. (see figure below)
For each of the four different dideoxynucleotides, we tag them with different fluorescent molecules where the colours of fluorescence when excited by light depends on the type of dideoxynucleotides. Therefore we may follow the procedure mentioned above to prepare the DNA fragments ending in eithe A, T, C or G depending on the type of dideoxynucleotides and thus the colour of fluorescence.
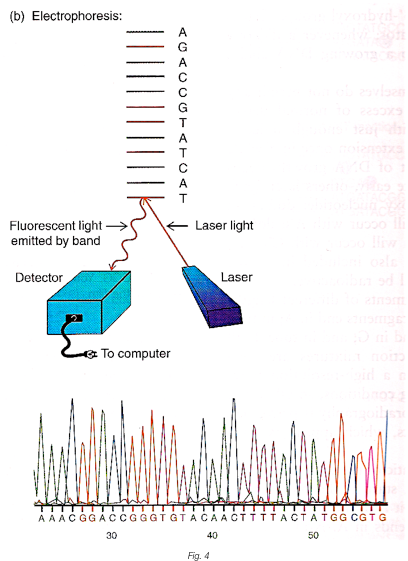
Then we may use gel electrophoresis to differentiate those DNA fragments from each other after denaturing them to single strands. Gel electrophoresis is simply allowing those electrically charged molecules to start from the same point and move through a thin column filled with gel, under a uniform electric field parallel to the column. The smaller the fragment is, the faster it will move through the column.
After this, the column is filled with DNA fragments across the gel comming in an order, this order is simply the sequence of the initial DNA being sequenced. To detect the sequence, we may use a lasor to excite each of the fluorescent molecules in series and in the meanwhile, record the colour of fluorescence being emitted from the molecule with a light detector. (see figure below)
(To learn further, go check out this short clip animating the process)
References
Figures obtained from Weaver, R. F.: Molecular Biology (3rd Ed.) (2005).