Growing genes is an essential process of genetic engineering. Producing required genes in a large quantity would allow researchers to acquire significant amount of raw materials required for development. The larger the amount of genes, the more probable it is going to be successful in engineering. Because for the designs we wanted there are always a large proportion of unwanted byproducts, to synthesise a considerable amount of final products we need to maximise the quantity of materials for developement.
Before we begin, we must have a perception of how a DNA (deoxyribonucleic acid) is structured. A DNA molecule is formed from 2 strands of long molecule chains called nucleotides. On each nucleotides there are four different types of nucleobases known as adenine, guanine, thymine and cytosine, or A, G, T and C for short. The 2 strands goes in opposite direction to each other, each with a sequence of nucleobases. Every nucleobases on a strand binds to another one on the other strand, where adenine only binds to thymine, and guanine only binds to cytosine and vice versa.
We start with the chemically synthesised primers (also called DNA oligonucleotides) on hand, which is namely short nucleic acid polymers1, acting as templates for our target DNA segment. Each of them contains the specific complementary bases of our genes of interest with the base sequences of the target DNA, either obtained from DNA sequencing which we will discuss in the next part, or entries in any of the various existing information libraries. In the same way DNA strands binds together, primers will also bind to DNA strands when possible.
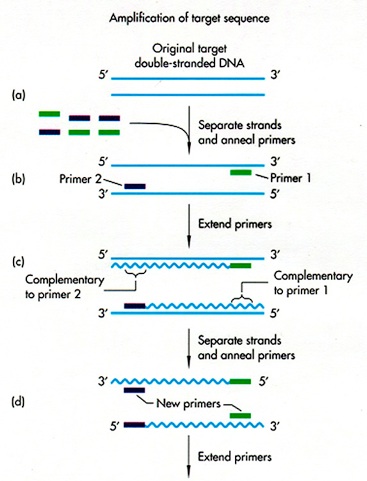
Then it undergoes what is called the polymerase chain reaction (PCR) process with reagents including the oligonucleotides we prepared, the DNA molecules containing our target, and other necessary reagents.
First, we heats the reagents to cause DNA molecules to ‘denature’ into single strands. Then the temperature of the mixture is lowered to allow primers to bind, or in Biology terms ‘to anneal’, to those single strand DNA molecules, as a starting point for the complementary strand. Finally polymerase will ‘extend’ / ‘elongate’ the primer, starting from the position of the primer, going through the DNA to match up the existing template strand with complementary base pairs, consuming materials known as the nucleotides (the bases of DNA molecules), to form a double helix with the existing strand. We can visualise the process of elongation as an analogy of a zipper on a jacket. The zipper slider works as the polymerase, while the existing template strand works as one of the tapes; the primer works as the retainer box on a zipper, defines the starting position of elongation; and finally, we construct the other side from base materials, the nucleotides, to perfectly match the shape of the template tape as the zipper slider moves along.
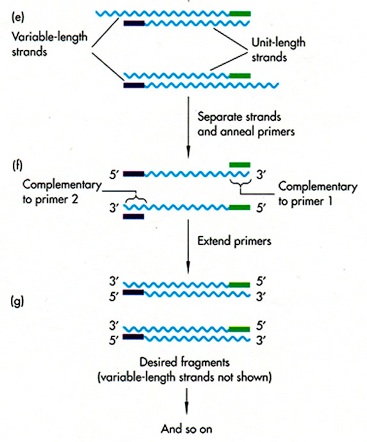
These three steps above double the number of the DNA fragments including our genes of interest and they are allowed to be repeated many times. In consequence we will observe an exponential growth, or ‘amplification’, of the DNA fragments required. (see figure below)